- Home
- About Us
- Our Work
- International Cooperation
- News & Events
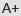



Progress on editing rice yield-related QTLs in different rice varieties
Oct 20th, 2016
Grain yield is the most important and complex trait for genetic improvement in crops; it is known to be controlled by a number of genes known as quantitative trait loci (QTLs). In the past decade, many yield-contributing QTLs have been identified in crops. However, it remains unclear whether those QTLs confer the same yield performance in different genetic backgrounds.
In the study, researchers from China National Rice Research Institute (CNRRI) and Yangzhou University performed CRISPR/Cas9-mediated QTL editing (GS3 and Gn1a) in five widely-cultivated rice varieties and obtained 10 novel genotypes. The results revealed that in all genetic back grounds the grain lengths of the gs3 and gs3gn1a mutants were increased compared to the WT. In addition, the grain number was also enhanced when Gn1a was mutated. Interestingly, the authors found that 7 of the 10 novel genotypes displayed decreased grain yields, while only 3 genotypes showed higher grain yields than the WT.
Grain yield of rice is determined by three factors: the number of effective tillers or panicles per plant, the number of grains per panicle, and the grain weight. To elucidate the underlying causes of the changes in grain yield, the researchers divided tillers into two categories depending upon the grain number: major tillers and minor tillers, and found that the number of major tillers is responsible for the change of grain yield. It is well known that grain yield per plant does not necessarily accord with population yield in large-scale field trials. Thus, compatible field management, such as rational dense planting, may be required for the application of all those genotypes in the future.
This work was supported by the Agricultural Science and Technology Innovation Program. The research finding has been published in Journal of Integrative Plant Biology online on September 18th 2016 (DOI: 10.1111/jipb.12501). More details are available on the links bellow:

Figure 1. Targeted mutation of GS3 and Gn1a led to diverse yield performance in different rice varieties.
· Genomic architecture of heterosis for yield traits in rice
· Progress on Replication Mechanism of DNA Polymerase in Insect Baculovirus
· Progress on Molecular Mechanism of Leaf Senescence via NAD pathway in Rice
· Range of CRISPR/Cas9 Genome Editing is Expanded in Rice
· CAAS Researchers Reveal a Positive Regulator of Rice Defense Response
-
CNRRI Today
-
About Us
-
Our Work
-
International Cooperation
-
News & Events