- Home
- About Us
- Our Work
- International Cooperation
- News & Events
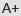



OsCUL3a Negatively Regulates Cell Death and Immunity by Degrading OsNPR1 in Rice
February 9th, 2017
The ubiquitin/proteasome pathway is a major system for the selective degradation of proteins in eukaryotes. Previous studies in plants have demonstrated that the ubiquitin/proteasome pathway is an essential component of the regulatory networks controlling many important cellular processes, including growth and development, as well as defense responses. Among the three major enzymes in this pathway, the E3 ubiquitin ligases are responsible for controlling the specificity of target protein recognition and degradation via the 26S proteasome. A prominent subset of the E3 ligases are the Cullin-RING E3 ligases (CRLs), in which the Cullin3 (CUL3)protein is known to function in the regulation of PCD in animals. In plants, the transcription coactivator NPR1 (NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1) is a master regulator of both basal and systemic acquired resistance (SAR). OsNPR1 over-expression rice plants have enhanced resistance to the bacterial pathogen Xanthomonas oryzae pv. Oryzae (Xoo), elevated expression levels of pathogenesis-related (PR) genes, and a benzothiadiazole-mediated cell death phenotype. Although OsNPR1 has been identified as a positive regulator of resistance against bacterial pathogens, its protein turnover and molecular mechanism remain largely unknown.
Recently, researchers from China National Rice Research Institute (CNRRI), Institute of Plant Protection, Chinese Academy of Agricultural Sciences, The Ohio State University, Zhejiang Key Laboratory of Super Rice Research and Henan Agricultural University identified a rice lesion mimic mutant, oscul3a, in a mutant collection. Their research findings show mutant oscul3a displays a significant increase in the accumulation of flg22- and chitin-induced reactive oxygen species, and in pathogenesis-related gene expression as well as resistance to Magnaporthe oryzae and Xanthomonas oryzae pv. oryzae. They cloned the OsCUL3a gene via a map-based strategy and found that the lesion mimic phenotype of oscul3a is associated with the early termination of OsCUL3a protein. Interaction assays showed that OsCUL3a interacts with both OsRBX1a and OsRBX1b to form a multi-subunit CRL in rice. Strikingly, OsCUL3a interacts with and degrades OsNPR1, which acts as a positive regulator of cell death in rice. Accumulation of OsNPR1 protein is greater in the oscul3a mutant than in the wild type. Furthermore, the oscul3a osnpr1 double mutant does not exhibit the lesion mimic phenotype of the oscul3a mutant. The result demonstrates that OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice.
This work was supported by was supported by the National Key Transgenic Program (2016ZX08001002), the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-CNRRI), the National Natural Science Foundation of China (#31571944), and the China Association for Science and Technology. The research finding has been published in Plant Cell online on January 18, 2016 (DOI: 10.1105/tpc.16.00650). More details are available on the links bellow:

OsCUL3a assembles with OsRBX1 and an unknown BTB domain containing protein to form an E3 ligase complex that promotes the degradation of OsNPR1 via the 26S proteasome system. OsNPR1 highly accumulates in oscul3a mutant plants, resulting in cell death and immunity.

(A) Delimitation of the candidate genomic region of lm-ZH. a. Preliminary mapping of the lm-ZH gene using 94 recessive F2 plants with simple sequence repeat markers. The numbers under the linkage map represent the number of recombinants. b. Fine mapping of the lm-ZH locus with the InDel markers. The numbers under the linkage map represent the number of recombinants. c. The arrows denote the candidate (ORFs) within the genomic region between the ZN36 and ZN9 markers. d. Structure of the lm-ZH gene and the mutation site. The line represents the intron; black boxes represent the exons.
(B) Protein alignment between wild-type OsCUL3a and the deduced 69 amino acid product in lm-ZH.
(C) Phenotype of 2-month-old complemented T1 plants grown in the field conditions. pOsCUL3a complementation plants were developed by transforming the whole genomic fragment of OsCUL3a into the lm-ZH mutant background. pOsCUL3a plants were photographed at 60 dps.
(D) Leaves (second from the top) of lm-ZH mutant and pOsCUL3a plants at 60 dps.
(E) DAB staining of the lm-ZH mutant and pOsCUL3a leaves shown in (D).
(F) Punch inoculation of lm-ZH mutant and pOsCUL3a plants with the compatible M. oryzae isolate RB22. Leaves were photographed at 12 dpi.
(G) Lesion area of the inoculated leaves of lm-ZH mutant and pOsCUL3a plants at 12 dpi. Error bar represent the SEM, n=3. (t-test, *P<0.05).
(H) Fungal biomass of the inoculated leaves of lm-ZH mutant and pOsCUL3a plants at 12 dpi. Error bar represent SEM, n=3. (t-test, *P<0.05).
-
CNRRI Today
-
About Us
-
Our Work
-
International Cooperation
-
News & Events